Research focus
Plant cells are capable of producing a variety of secondary metabolites (specialized metabolites). These small organic molecules allow plants to cope with various types of stresses and also have biological activities of high interest to human, e.g. artemisinin, an anti-malarial compound produced by Artemisia annua plants. Yet, plant secondary metabolic mechanism is still scarcely exploited, mainly because of the incomplete molecular insight into plant secondary metabolism. Our lab, using medicinal plants (Artemisia annua, Catharanthus roseus) as research objects via cutting-edge functional genomics tools, in combination with reverse genetics screenings, aims to decipher the molecular mechanisms regulating plant secondary metabolites biosynthesis.
We have created a functional genomics based technology platform that enables comprehensive investigations and large-scale gene discovery in plant secondary metabolism and trichome development. Our platform is built on the integration of genome, transcriptome, proteome and metabolome profiling data. Based on these data, a large numbers of useful genes are screened out that allow us to increase our fundamental understanding of plant secondary metabolism, which facilitates the production of valuable plant-derived metabolites in medicinal plants.
1)Research and Development on Artemisia annua and Artemisinin
- Omics Study on Artemisia annua
Artemisia annua, commonly known as sweet wormwood or Qinghao, is a shrub native to China and has long been used for medicinal purposes. A. annua is now cultivated globally as the only natural source of a potent anti-malarial compound, artemisinin. Here, we report a high-quality draft assembly of the 1.74-gigabase genome of A. annua, which is highly heterozygous, rich in repetitive sequences, and contains 63 226 protein-coding genes, one of the largest numbers among the sequenced plant species. We further revealed by transcriptome profiling that A. annua has evolved the sophisticated transcriptional regulatory networks underlying artemisinin biosynthesis. Based on comprehensive genomic and transcriptomic analyses we generated transgenic A. annua lines producing high levels of artemisinin, which are now ready for large-scale production and thereby will help meet the challenge of increasing global demand of artemisinin.
Proteomics and metabolomic analysis platform have been established to comprehensively analyze artemisinin biosynthesis pathway by combining proteome and metabolomic data.
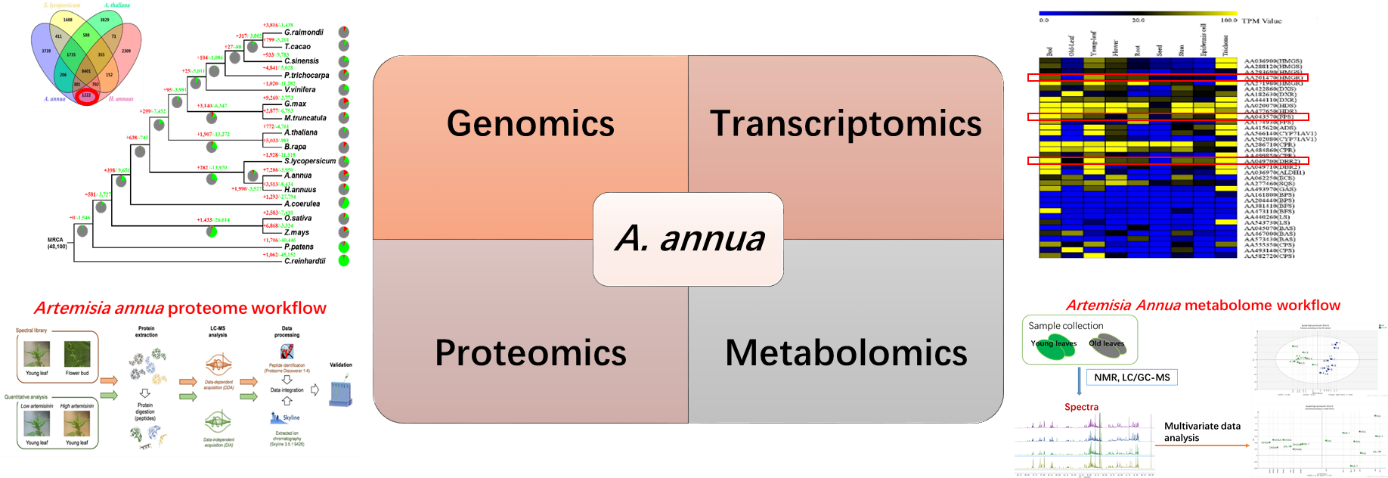
Fig 1 Omics Study on Artemisia annua
- Research and development of natural bioactive compounds
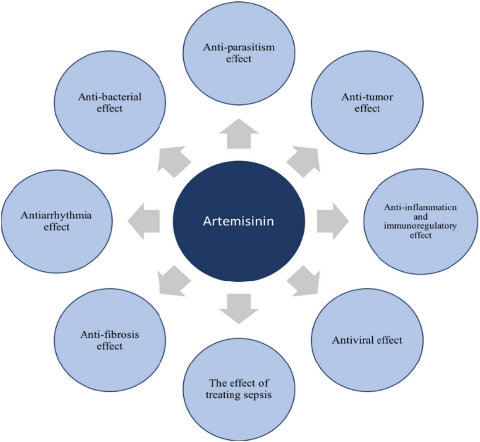
In 2016, cell published that artemisinin can be used to treat diabetes; in 2016, nature chemical biology published artemisinin in the treatment of tuberculosis; in 2017, dihydroartemisinin in the treatment of lupus erythematosus entered the phase II clinical trial. The center took the lead in studying and exploring the efficacy of artemisinin in reducing blood lipid and treating fatty liver and hepatitis B.
2)Plant Synthetic Biology
Marchantia polymorpha (liverwort), A. annua, tobacco and tomato are used as the matrix plants for synthetic biology study in our lab. We have successful produced high-level patchoulol in A. annua using synthetic biology tools. Furthermore, we have collected various liverwort species, and established efficient culture and transformation systems. Currently we are working on producing high-valued biomolecules such as santalol, β-elemene and vinblastine in liverwort, A. annua, tobacco and tomato.
3)Metabolic regulation and metabolic engineering of plant natural porducts
Artemisinin biosynthesis and regulation:Based on our multiple A. annua organs or tissues transcriptome sequencing database, by using gene co-expression analysis strategy, we characterized transcription factors involved in artemisinin biosynthesis and established related transcriptional regulation network.
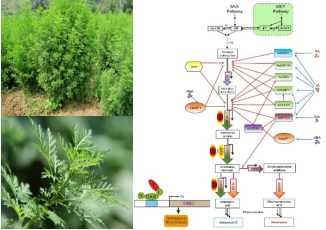
Figure 3. The transcriptional regulation network of artemisinin in A. annua
Trichome development:Trichomes especially glandular trichomes, regarded as bio-factories, have a unique capacity for biosynthesis and storage of secondary metabolites. Only limited trichome genomic information is available. By using Laser capture microdissection technology, we separated A. annua glandular and non-glandular trichome cells and generated transcriptome sequencing databases. By screening these databases, we identified a number of genes that regulate trichome initiation.

Figure 4. Morphology of trichomes and molecular mechanism of glandular trichome initiation in A. annua
Metabonomic profiling and MIAs pathway regulation of Catharanthus roseus: As the natural resource for antitumor agents vinblastine and vincristine, C. roseus is highly valued and has been studied extensively as a model of medicinal plants. Monoterpenoid indole alkaloids (MIAs) biosynthesis is complicated and highly regulated. Our lab is the only one that has established stable transformation of C. roseus via Agrobacterium-mediated method. Combined with 1H-NMR based metabolomics, we could explore this model medicinal plant more efficiently.
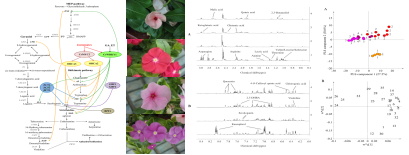
Figure 5 MIAs pathway and metabolic profiling of C. roseus plants with different flower colors

